Welcome Polygenically Screened Babies
Another thing I missed during my hiatus last year: the birth of the first polygenically-screened baby.
[conflict of interest notice:LifeView, the company that handled the screening, was co-founded by Steve Hsu. I’ve known Steve for many years now, he is very nice to me, always patiently answers my genetics questions, and sometimes comes to SSC/ACX meetups]
During in vitro fertilization, a woman takes drugs that make her produce lots of eggs. Doctors extract the eggs and fertilize them with sperm from a partner or donor, producing lots of embryos. Hopefully at least one of the embryos looks healthy, and then the doctors implant it in the woman or a surrogate parent.
For a while now, if the process produces enough embryos, doctors have used some simple low-tech genetic tests to choose the healthiest. For example, they might look for Down syndrome or other obvious chromosomal abnormalities, or for very severe monogenic diseases like sickle cell anemia. All of this is routine.
But most interesting traits aren’t chromosomal abnormalities or single genes. They’re polygenic, ie controlled by the interaction of thousands of genes. You need to record basically the entire genome and have a good idea what all of it is doing before you can predict these. Right now we’re at an inflection point where we can sort of do this for a few traits, and some companies are starting to apply this to the embryo selection process.
I often have patients ask me something like: “I have a history of schizophrenia in my family. I’m really concerned my kid might get schizophrenia. What can I do to prevent this?” Right now I don’t have a lot of answers, besides just staying generally healthy during pregnancy and making sure the kid has a healthy upbringing. But with polygenic screening, you start to get more options. You can IVF lots of embryos, test all of them for genetic schizophrenia risk, and implant whichever one gets the lowest score. How much does that help? LifeView, the pioneering polygenic screening company, has some helpful calculators:
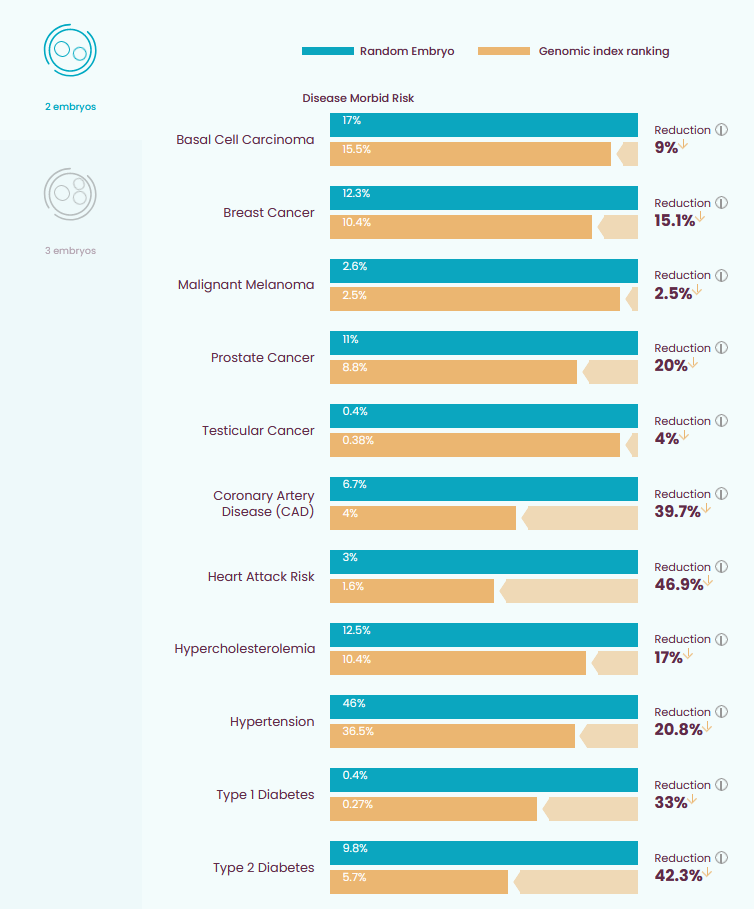
 It won’t let me calculate this one for more than two embryos, or for people with a family history of the disorder, but with enough embryos probably you could cut your risk by half or more.
It won’t let me calculate this one for more than two embryos, or for people with a family history of the disorder, but with enough embryos probably you could cut your risk by half or more.
What if you have a family history of multiple conditions, like schizophrenia and breast cancer? Wouldn’t that mean decreasing returns on selection? Unless by coincidence the embryo with the lowest schizophrenia risk also has the lowest breast cancer risk, you have to choose to minimize one or the other, right?
Sort of. You can also just try to minimize serious disease. This is the idea behind genomic indexing, which calculates an embryo’s risk of every disease we know how to screen for, weights them by how likely they are and how bad they are, and tries to pick the embryo with the best chance of an overall healthy life. So for example, if Embryo 1 has high risk of breast cancer but low risk of high blood pressure, and Embryo 2 is the opposite, then the algorithm knows that cancer is worse than high blood pressure, and will select Embryo 2. Here’s what you can do with genomic indexing:
Are these levels of reductions “worth it”? I think the company has a pretty good pitch, which is - if you’re doing IVF, you have to choose an embryo anyway. If you don’t screen, you’re going to choose one at random. So why not instead choose one in a way that halves your future kid’s risk of serious diseases? Of course, “worth it” depends on the price, which the company is kind of coy about, but an earlier version seems to have cost ~$1400 plus some extra per embryo, which is a fraction of overall IVF costs and pretty cheap for a US medical procedure
You’ll notice that all the traits being measured here are pretty serious medical conditions. In theory, you’re not supposed to use polygenic screening to produce designer babies. What about in practice? Screening companies will give you the raw data if you ask for it, so if you want to screen for an embryo with green eyes, all you need to do is find some third party algorithm that can screen genomes to figure out the baby’s eye color and plug in your data. Does anything like this exist? I don’t think so, but I think it would be trivial for a genetics PhD student to make.
[edit: existing polygenic screening companies might not read enough genes to do this. Although it would be easy to offer a more complete service that reads most genes, banning the more complete version might be one way regulators could prevent this otherwise hard-to-prevent thing.]
What about IQ? There are definitely scientists who have figured out how to do polygenic analyses to predict a modest amount of variation in IQ, though I don’t know if their algorithms are public, and they’re certainly not convenient for amateurs to use. If you had them, would they work? Gwern has done some calculations and finds that with ten embryos (a near-best-case scenario of what you’re likely to get from egg extraction) and modern (as of 2016) polygenic scoring technology, you could get on average +3 IQ points by implanting the smartest. If polygenic scoring technology reached the limits of its potential (might happen within a decade or two) you could get +9 IQ points. Embryos from the same parents only vary a certain amount in IQ, and about half of IQ variation is non-genetic, so you can’t work miracles with this (if you want to know how to work miracles, read the rest of Gwern’s article).
 Prediction markets say to wait fifteen years
Prediction markets say to wait fifteen years
Getting back to reality - the first polygenically screened baby was born last year to a family with a history of breast cancer, which screening + selection can make less likely. Her name was Aurea, meaning “dawn” - which may or may not be a coincidence.
 And either she looks like this, or else this was a stock photo of a baby someone used on the article about her. She shares an unusual last name with a commenter on this blog, so maybe we’ll get some updates!
And either she looks like this, or else this was a stock photo of a baby someone used on the article about her. She shares an unusual last name with a commenter on this blog, so maybe we’ll get some updates!